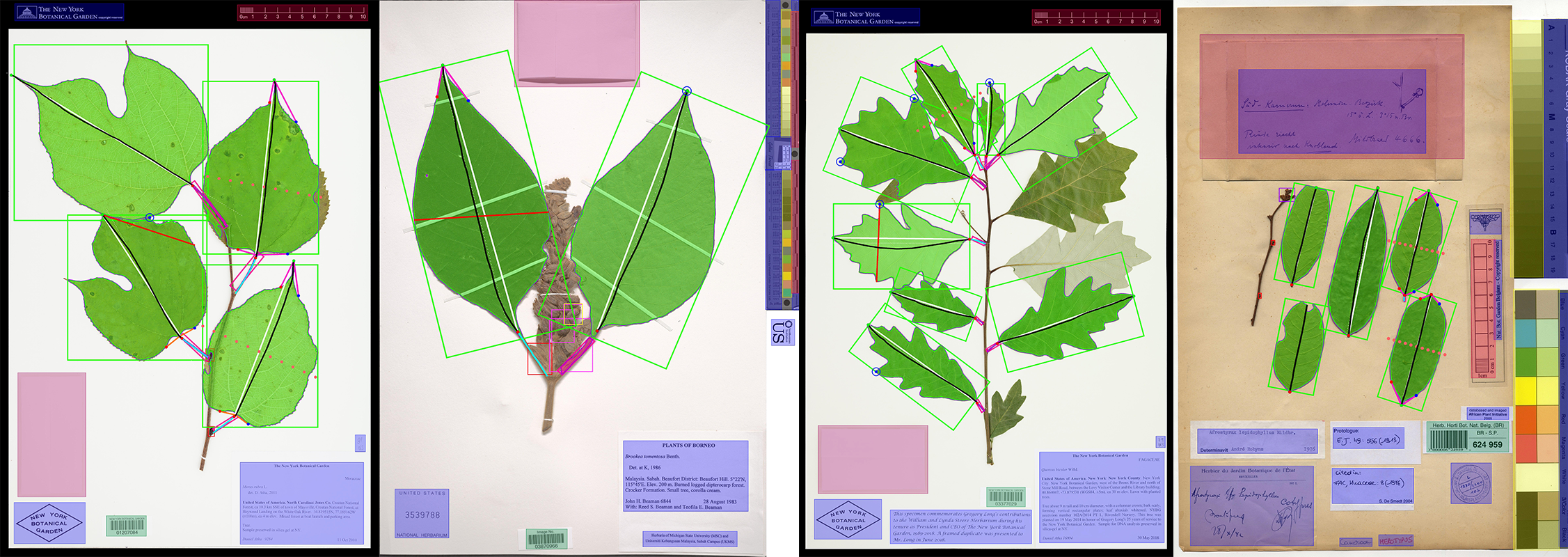
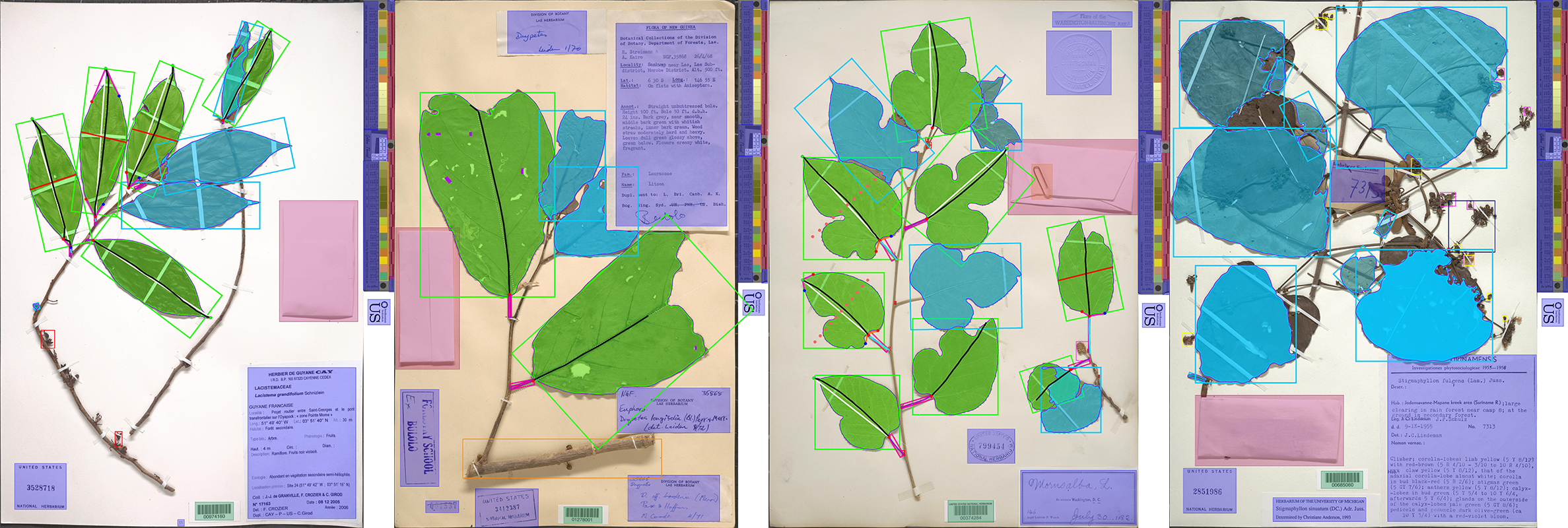
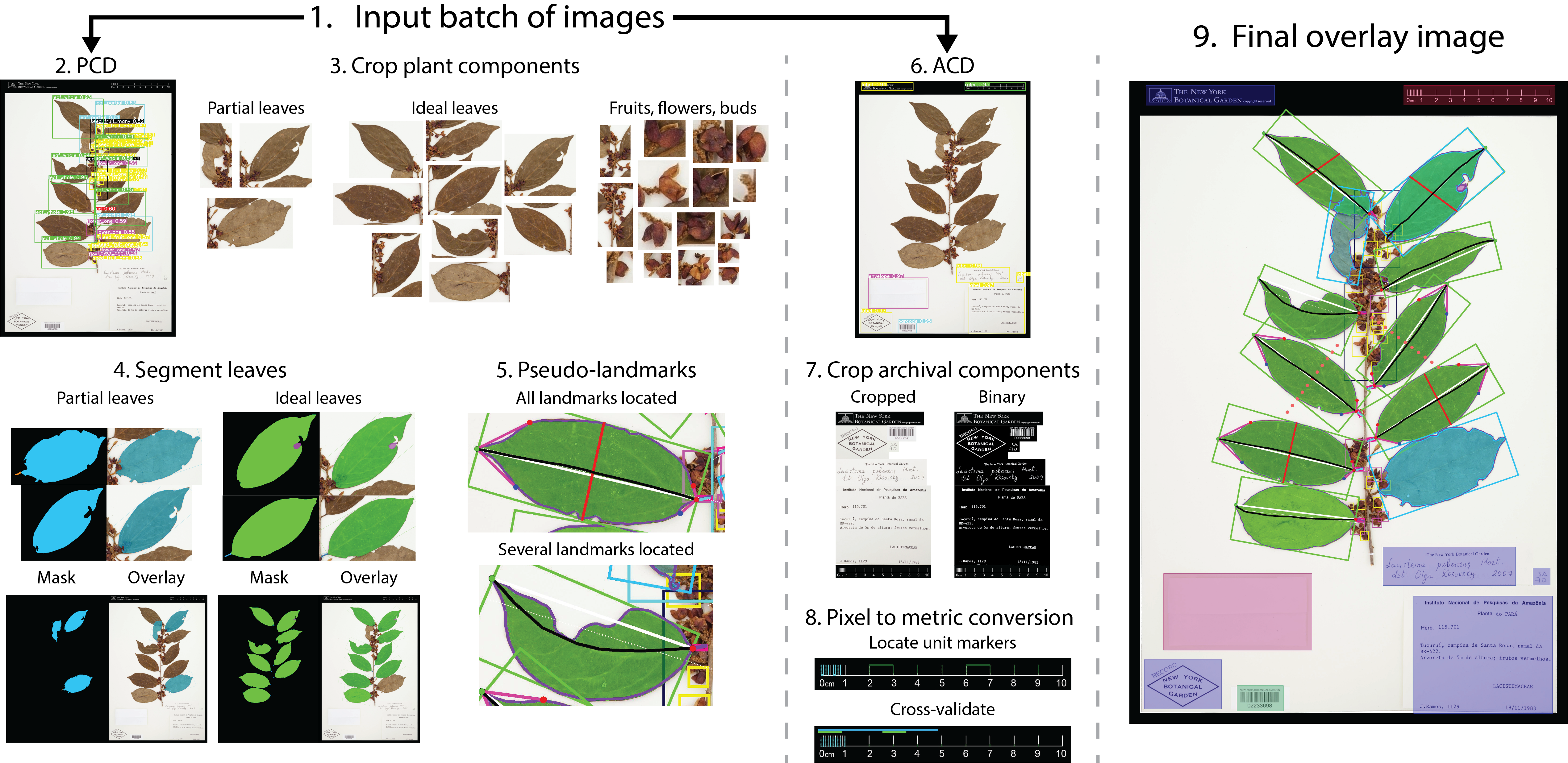
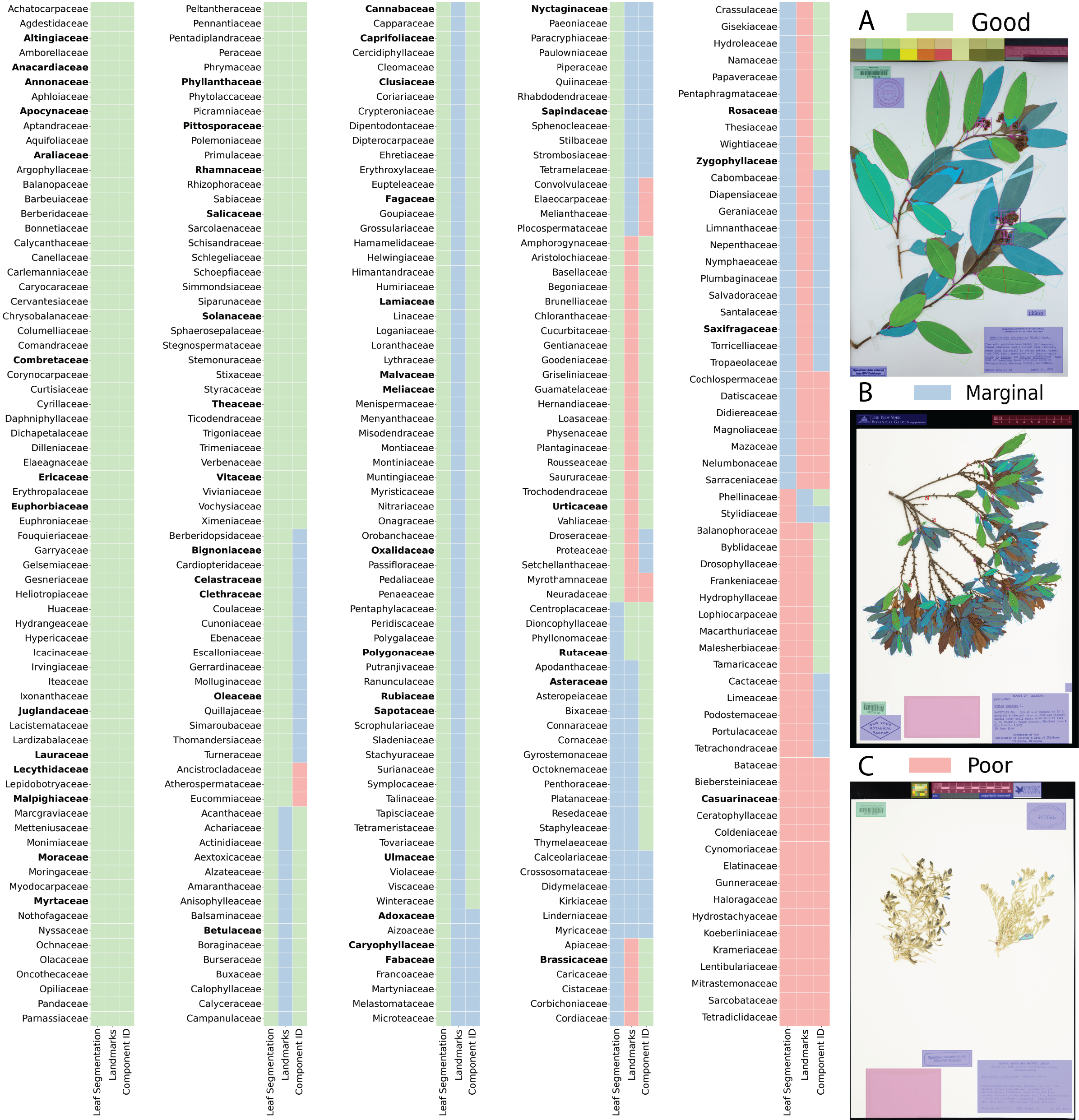
LeafMachine2
Post-Publication Updates!
March 22, 2025
New VoucherVisionGO API!
We're excited to announce the release of the VoucherVisionGO API, a simplified way to access VoucherVision's AI transcription capabilities for museum specimens. This API provides the best OCR/LLM combination for AI transcription using Google Gemini models.
The API is designed for integration with your projects and portals (Symbiota, Specify, etc.) and is simpler to use than the full VoucherVision implementation.
The VoucherVisionGO API offers:
- Single image file upload processing
- Single image URL processing
- Batch processing of URLs from text or CSV files
- Batch processing of folders containing images
- Results delivered in CSV or JSON format
September 5, 2024
This update includes:
- Very important update, LM2 finally stores data in a SQLite database, which makes exporting and manipulating data much easier.
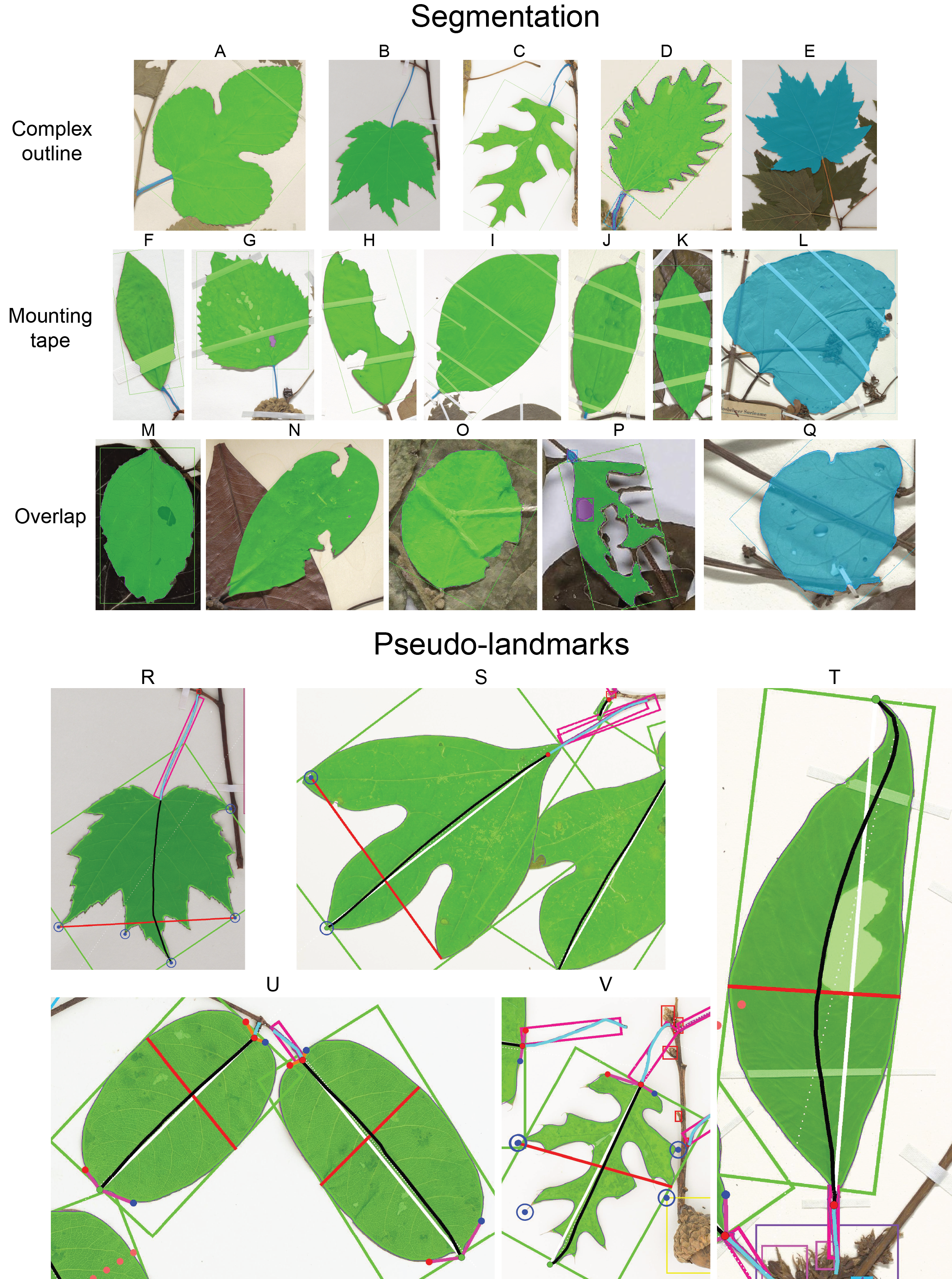
- Updated summary image (attached) with the keypoints plotted instead of the pseudolandmarks. Both options are available, but the keypoints are actually used to orient the leaf masks.
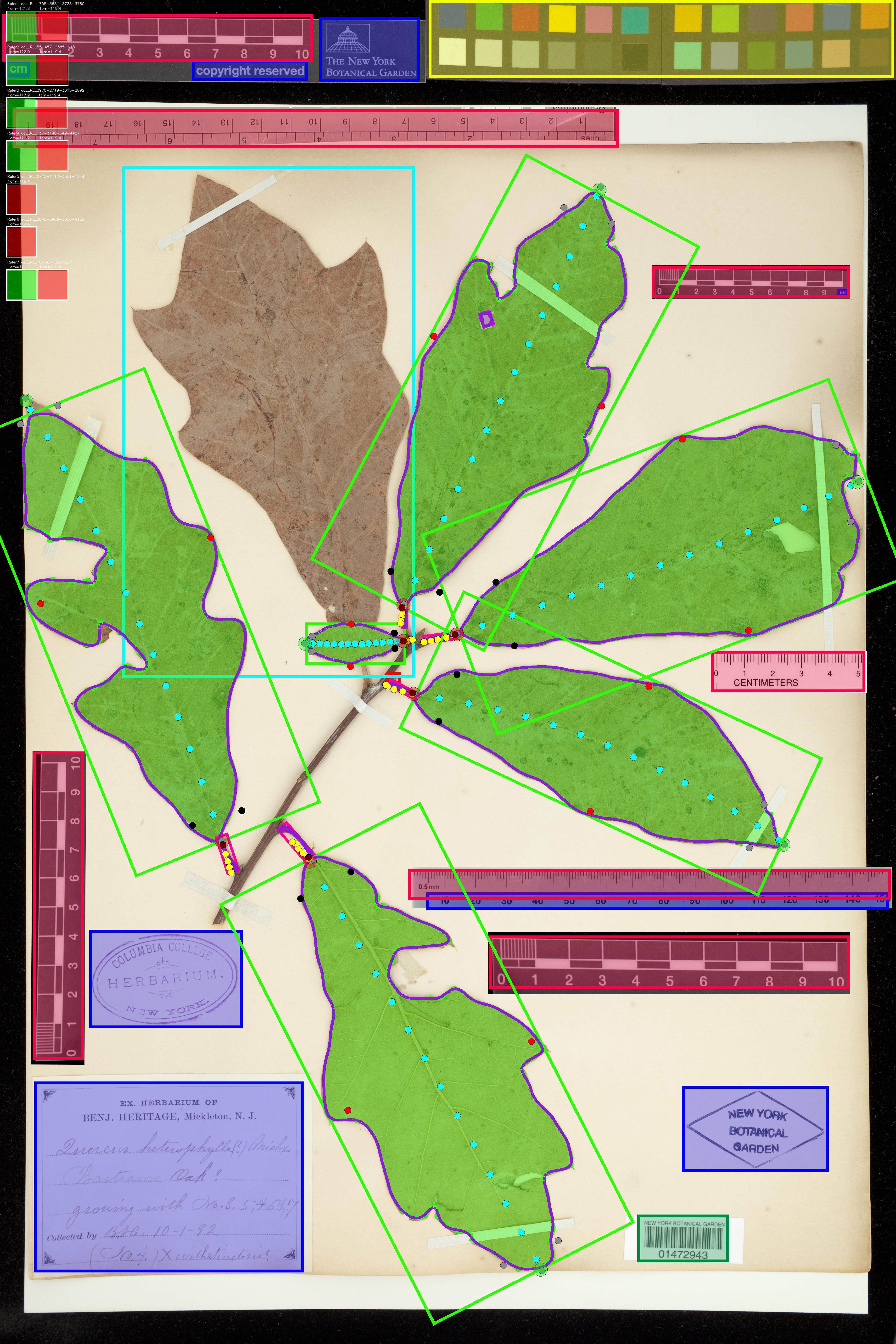
New Summary Image: Click the image to see it in more detail. Keypoints instead of pseudo-landmarks. Also, in the top left I now include a pixel-to-metric verification overlay. For each ruler in the image there can be a green square (calculated mean conversion factor) and a red square (predicted conversion factor). The predicted CF is based only on a linear projection of the image resolution and assumes that the input image is a standard herbarium specimen photographed on an 11x17 page. The predicted CF is only used if the calcualted CF is not available. This demo image is just to stress test by adding lots of different rulers.
May 12, 2024 (v-2-3)
This update includes:
- A new leaf segmentation algorithm trained on 3x more training images. It was also trained longer; it should produce more refined leaf masks.
- You can use LeafMachine2 to censor any archival component (rulers, labels, colorcards, etc.) with the color of your choice. Can be run separately from the rest of LM2 using CensorArchivalComponents.py and CensorArchivalComponents.yaml files. This is helpful for classification tasks when you don't want the model to be able to read the label text or link certain items with a certain class. An example image is below.
- I added a simple phenology detection tool. LM2 counts all of the detections from the PCD as a proxy for phenology. This runs by default with LM2, or you can run just the phenology detector via the DetectPhenology.py and DetectPhenology.yaml files. Running it seprately from the rest of LM2 is very efficient; processing 120,000 images only takes about 4 hours.
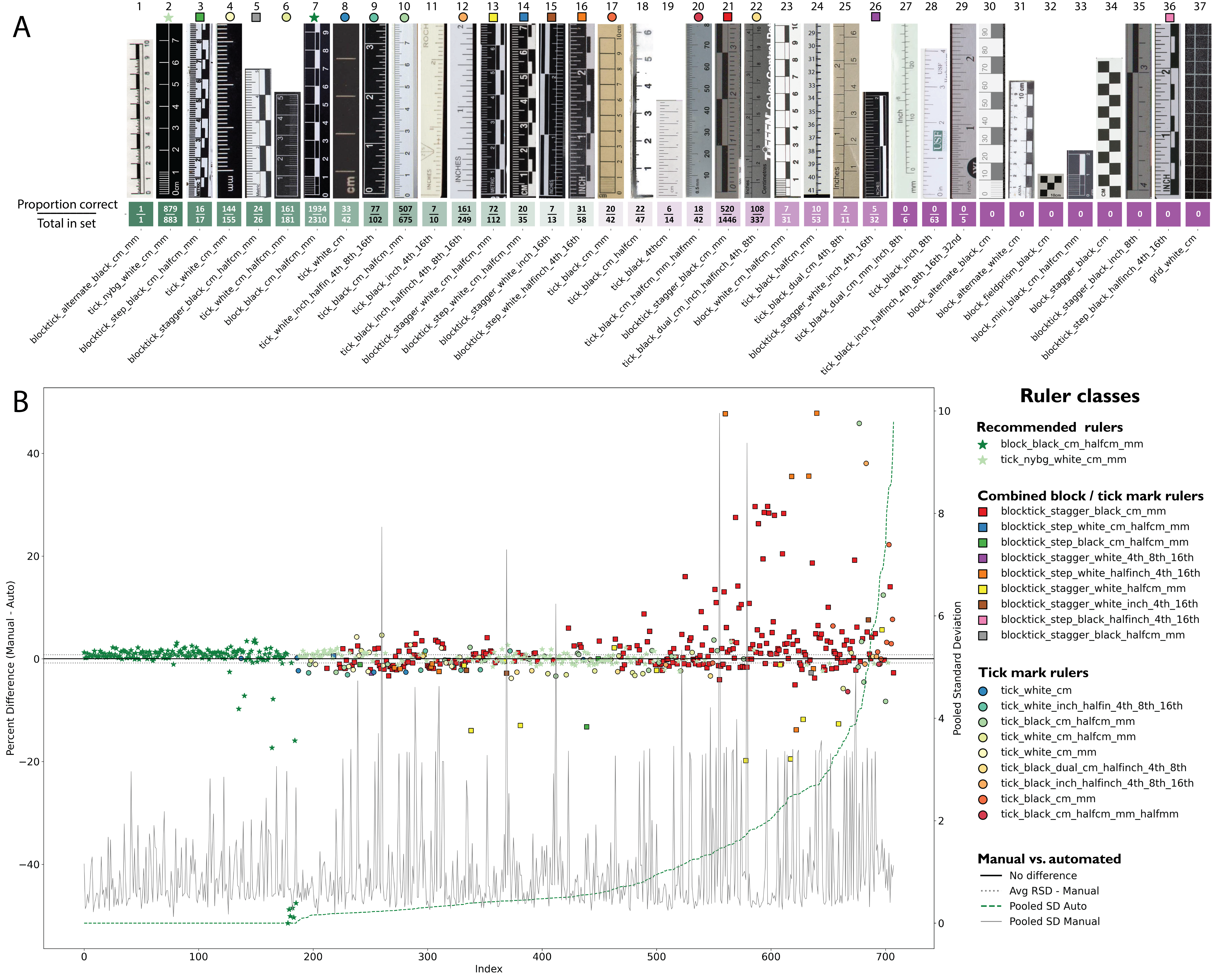
- I significantly improved the pixel to metric conversion performance. Now we can predict the conversion factor (CF) based on the image resolution, which then helps the LM2 algorithms detect the true CF's signature, cross-validating available units for each ruler. The new predicted CF acts as a baseline; if the algorithmically determined CF falls within 10% of the predicted CF, then we have extremely high confidence that the calculated CF is actually correct. For a 25 MP image from NYBG, the difference between the predicted CF and the True CF is less than the width of one of the ruler's tick marks. Note: this only works with standard herbarium images. If you are using LM2 with custom images, your mileage may vary.
- I reprocessed the D-3FAM dataset with this release (default settings, PCD set to 70%) and uploaded all output files to Zenodo. The original Zenodo repo only included PDFs of the summary images, but this includes everything. Link to all D-3FAM output files.

Censored Labels: You can pick the color to use to censor the labels (or any of the ACD classes). Here I censored everything. If you are training an image classifier, you will want to do this. ML models really like to focus on archival differences between images, since they are more prevalent and obvious patterns than the differences within the actual plant material.
March 25, 2024 (v-2-2):
This update includes:
We have released a new and improved Plant Component Detector (PCD)! This is the YOLOv5 object detection algorithm that is responsible for locating leaves, flowers, fruit, roots, etc.
What changed? Since publication we have quadrupled the size of our training dataset. The new PCD is trained on 20,000+ full resolution images and 600,000+ groundtruth bounding box annotations. In the paper we describe a few situations where the original PCD struggled. There was a small-leaf bias, so the new training dataset adds more large-leaf taxa including:
- Alismataceae Sagittaria
- Araceae Monstera
- Sapindaceae Acer
- Fagaceae Quercus
- Bignoniaceae Catalpa
- Phellinaceae Phelline
- Piperaceae Piper
- Platanaceae Platanus
- Melastomataceae Blakea
- Rubiaceae Posoqueria
- Bixaceae
- Pentaphragmataceae
- Torricelliaceae
- Aponogetonaceae Aponogeton
- Euphorbiaceae Macaranga
The new training dataset also includes the following images to improve generalizability across taxa and scenarios:
- 1,100+ bryophytes from the Oregon State University herbarium, courtesy of Dr. James Mickley.
- 9,000+ images from the LeafSnap dataset to bolster performance for single-leaf images. We did not use the LeafSnap provided segmentation masks. Instead, we manually segmented the RGB images to conform to our class definitions. Please see LeafSnap.com.
- 500+ field images (using FieldPrism photogrammetric templates), courtesy of Dr. Chuck Cannon, Claire Henley, and Arnan Pawawongsak, from the Morton Arboretum.
The new PCD is called LeafPriority and is optimized for locating woody perennial associated components including leaves, flowers, fruits, roots, etc., but has been retrained to handle the 'specimen' class in a different way. The 'specimen' class is designed to detect plant material when typical broad leaves are not present, like a bryophyte image. The new PCD improves all downstream machine learning components of LeafMachine2 (segmentation and landmarking) and also improves the SpecimenCrop feature, which is designed to automate the tedious process of removing whitespace from specimen images.
What did we learn? For LeafPriority training we removed all groundtruth bounding boxes for the 'specimen' class if whole or partial leaves were also present in the image. So while the original dataset contained more than 25,000 groundtruth examples of the 'specimen' class, LeafPriority was trained on only 2,000 examples of the 'specimen' class. Since the 'specimen' class originally encompassed all other plant classes, it erroneously linked large dimension bounding boxes (bounding boxes that occupy most of the image) only with the 'specimen' class, making it much less likely to correctly identify very large leaves or leaves that occupy most of the field of view, stemming from the YOLOv5 model's reliance on anchor bounding boxes for anticipating object locations.
How can I use the new model? The new model will be downloaded by default with new installations of LeafMachine2. If you already have LeafMachine2 installed it should automatically download the new version once you run a `git pull` to update your local repo. If using the LeafMachine2 GUI, the new default is LeafPriority. If you manually edit the LeafMachine2.yaml file, we include instructions for selecting the new or old model.
What's next? We have 5x the number of leaf segmentations since publication, stay tuned for an updated version! We also have a pose detection model trained to identify landmarks, which also significantly improves unpon the existing landmark detection algorithm.

Comparing the old PCD to the new LeafPriority PCD: Large leaves are much more likely to be detected. With the old PCD, most of the training data were densely packed herbarium specimens, so the PCD actually worked better with dense oeverlapping specimens and struggled with isolated leaves. LeafPriority no longer places 'specimen' bounding boxes (the black boxes) around isolated leaves, significantly improving detection accuracy, as you can see in the Ginkgo biloba and Castanea dentata images. The bryophyte images show a huge improvement; LeafPriority only places a single 'specimen' box around the material, instead of the messy Original detection.